In recent years, it has been increasingly recognized that sharing original data enhances transparency, reproducibility, and scientific impact. This shift has prompted a growing international movement to make publicly funded research data accessible to all. At the same time, turning this goal into reality presents challenges for both researchers and infrastructure providers.
From a facility perspective, having data management plans (DMPs) in place is essential. Facilities often support a large number of users with diverse experimental workflows, instrument requirements, and levels of experience. As a result, consistent data organization becomes critical to effectively navigate large and heterogeneous datasets. Streamlined workflows help reduce confusion, prevent duplication of effort, and optimize storage use—avoiding the accumulation of unnecessary or redundant data.
From a researcher’s perspective, the idea of sharing data often raises concerns, particularly the fear of being scooped or losing control over unpublished results. It’s important to clarify: data does not need to be made publicly available the moment it is generated. Instead, the key is to ensure that data are well-organized and FAIR (i.e., Findable, Accessible, Interoperable, and Reusable) by the time of publication. This timing protects the scientific lead of the researcher while aligning with journal, funder, and institutional requirements.
Well-organized data provides clear benefits for the research community, but also the individual researcher:
- improved understanding of the data, especially when thoughtful organization is applied early in the experimental design
- greater scientific impact, as others build upon the published work
- reduced delays and stress at the point of submission, as data are already well-documented and publication-ready
- improved credibility and transparency, reducing the risk of results being challenged or misunderstood
- increases visibility and citation rates when datasets are discoverable and reusable
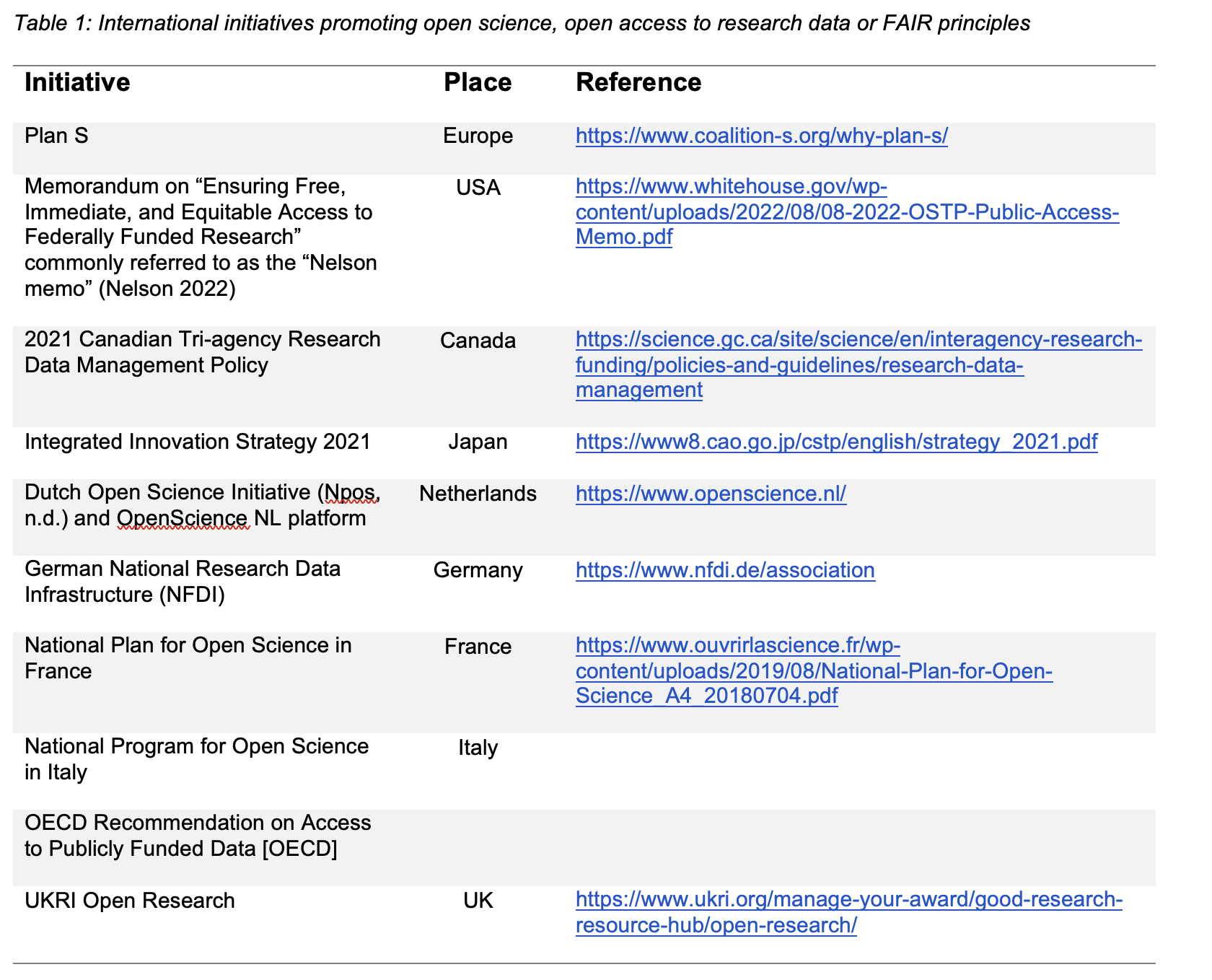
Various initiatives (listed in Table 1) have aimed at establishing or promoting open science, that is, making scientific results from publicly funded research accessible. Notably, the FAIR principles (Wilkinson et al., 2016) introduced a widely applicable framework for data management and data stewardship, , including of data that cannot be openly shared for valid reasons. Since then, many initiatives have promoted FAIR data by developing tools and standards to facilitate adherence to these principles across diverse scientific domains, including bioimaging. FAIR data enables reuse beyond the original context, boosting impact—as reflected, for example, by higher citation rates of publications associated with shared data—and thereby fostering trust and reliability for more effective and efficient scientific research.
As a result, data producers and managers are required to ensure that data are described properly and can be shared via institutional or disciplinary repositories.
Research data management (RDM) has thus become an integral mission of imaging facilities (https://doi.org/10.1002/jemt.22648; https://doi.org/10.1002/mrd.22538; https://doi.org/10.7171/3fc1f5fe.97057772; https://doi.org/10.1111/jmi.13317). RDM is often defined as all the activities in the research process that are not data analysis or processing. Specifically, research data management refers to the organization and handling of research data throughout the research data lifecycle from data management planning and data creation through to storage, backup, retention, archiving, destruction, access, preservation, curation, dissemination (publication, sharing, reuse), documentation and description of data, including the complementary algorithms, code, software and workflows that support research data management practices.